Equipment and products we offer
Novel formats for the comet assay
The traditional comet assay has 1 or 2 gels on a microscope slide, and there is a limit of about 40 gels per experiment. It is not suitable for large screening studies, and requires a high number of cells and large amount of drug or chemical to be tested. To optimise the method, we have developed two alternative formats, high- and medium- throughput. The new procedures save time at all stages and are suitable for further automation.
Medium throughput: 12-gels format
The medium throughput format involves 12 gels set on a microscope slide. A gasket with holes over the gel positions can be used, allowing differential treatment with chemicals, reagents or enzymes. Twenty slides can be run in one experiment, giving a total of 240 gels


High throughput: 48- or 96- gels format
High throughput format involves 48 or 96 gels set on sheets of films, which are cut to size and supported under tension by stainless steel frames. Four films can be run in one experiment, giving a total of up to 384 gels.


Tissue mincing device
Our tissue mincing unit is devised for rapid disaggregation of tissue for various experimental purposes including the comet assay. It is made of stainless steel and consists of a cylindrical tube ending with a grid. The unit is used with a standard syringe plunge and is eminently suitable for use in in vivo assays.


Reference standards
We provide human or rodent reference cells containing defined amounts of specific lesions:
- Strand breaks induced by X-irradiation, for basic comet assay
- Altered bases for enzyme-modified assay
- Negative controls
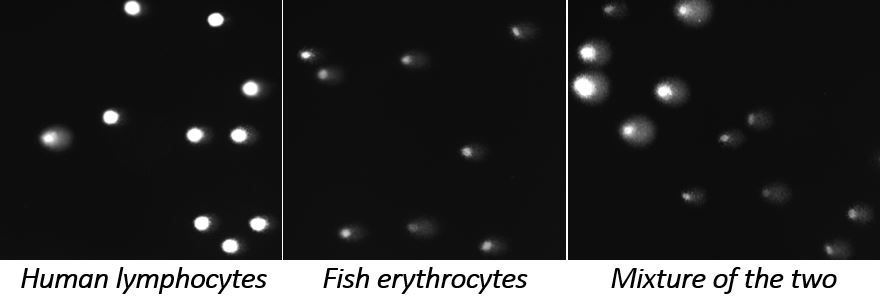
We also develop internal reference standards using cells from organisms with very different genome size compared with the experimental human or rodent cells. These reference standards can be present in the same gel with test cells, and are readily distinguished when they are imaged as comets.
Enzymes for specific base damage detection
We provide various repair enzymes for detection of specific DNA lesions, increasing the sensitivity and versatility of the comet assay as a genotoxicity test.
- Formamidopyrimidine DNA glycosylase (Fpg) acts as both N‐glycosylase and AP‐lyase; the N‐glycosylase activity releases damaged purines from double stranded (ds) DNA, generating an apurinic (AP site). The APlyase activity cleaves both 3’ and 5’ to the AP site, removing the AP site and leaving a 1‐base gap. Damaged bases recognized and removed by Fpg include 8‐oxoguanine, 8‐oxoadenine, fapy‐guanine, methyl‐fapyguanine, fapy‐adenine, aflatoxin B1‐fapy‐guanine, 5‐hydroxycytosine and 5‐hydroxyuracil.
- Endonuclease III (endoIII, Nth) acts as both N‐glycosylase and AP‐lyase. The N‐glycosylase activity releases damaged pyrimidines from ds DNA, generating an AP site. The AP‐lyase activity of the enzyme cleaves 3’ to the AP site leaving a 5’ phosphate and a 3’ ring opened sugar. Damaged bases recognized and removed by Endo III include urea, 5, 6 dihydroxythymine, thymine glycol, 5‐hydroxy‐5 methylhydantoin, uracil glycol, 6‐hydroxy‐5, 6‐dihdrothimine and methyltartronylurea.
- 8‐Oxoguanine glycosylase (hOGG1) is the eukaryotic counterpart of Fpg, but tends to be more specific. Damaged bases recognized and removed by hOGG1 include 8‐oxoguanine when base‐paired with cytosine, 8‐oxoadenine when base‐paired with cytosine, fapy‐guanine and methyl‐fapy‐guanine.
We can also provide enzymes with other substrate specificity and establish the protocols for including them in the comet assay.
References
- Shaposhnikov S, Azqueta A, Henriksson S, Meier S, Gaivão I, Brunborg G, Nilsson M, and Collins A (2010) Twelve-gel slide format optimised for comet assay and fluorescent in situ hybridisation. Toxicology Letters, 19,195:31-4. DOI: 10.1016/j.toxlet.2010.02.017.
- Gutzkow K, Langleite T, Meier S, Graupner A, Collins A, Brunborg G (2013) High throughput comet assay using 96 minigels. Mutagenesis 28, 333‐340. DOI: 10.1093/mutage/get012.
- Shaposhnikov S, Brunborg G, Azqueta A, Collins A (2013) Novel formats for the comet assay. Toxicology Letters: 221:S189. DOI: 10.1016/j.toxlet.2013.05.431.
- Brunborg G, Jackson P, Shaposhnikov S, Dahl H, Azqueta A, Collins A, Gutzkow K (2014) High throughput sample processing and automated scoring. Frontiers in Genetics 10/2014; 5:373. DOI: 10.3389/fgene.2014.00373.
- Brunborg G, Collins A, Graupner A, Gutzkow K, Olsen A-K (2015) Reference cells and ploidy in the comet assay. Frontiers in Genetics 6:61. DOI: 10.3389/fgene.2015.00061.
- Azqueta A, Shaposhnikov S, Collins A (2009) Detection of oxidized DNA using DNA repair enzymes. The Comet assay in toxicology. Edited by Dr Alok Dhawan and Professor Diana Anderson. RSC Publishing.
- Brunborg G, Collins A, Graupner A, Gutzkow K, Olsen A-K (2017) Corrigendum: Reference cells and ploidy in the comet assay. Frontiers in Genetics 8:4. DOI: 10.3389/fgene.2017.00004.
